Equipe DesCarTes
Groeneveld CS, Sanchez-Quiles V, Dufour F, Shi M, Dingli F, Nicolle R, Chapeaublanc E, Poullet P, Jeffery D, Krucker C, Maillé P, Vacherot F, Vordos D, Benhamou S, Lebret T, Micheau O, Zinovyev A, Loew D, Allory Y, de Reyniès A, Bernard-Pierrot I, Radvanyi F.
European Urology 2024, May;85(5):483-494.
Background
Molecular understanding of muscle-invasive (MIBC) and non–muscle-invasive (NMIBC) bladder cancer is currently based primarily on transcriptomic and genomic analyses.
Objective
To conduct proteogenomic analyses to gain insights into bladder cancer (BC) heterogeneity and identify underlying processes specific to tumor subgroups and therapeutic outcomes.
Design, setting, and participants
Proteomic data were obtained for 40 MIBC and 23 NMIBC cases for which transcriptomic and genomic data were already available. Four BC-derived cell lines harboring FGFR3 alterations were tested with interventions.
Intervention
Recombinant tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), second mitochondrial–derived activator of caspases mimetic (birinapant), pan-FGFR inhibitor (erdafitinib), and FGFR3 knockdown.
Outcome measurements and statistical analysis
Proteomic groups from unsupervised analyses (uPGs) were characterized using clinicopathological, proteomic, genomic, transcriptomic, and pathway enrichment analyses. Additional enrichment analyses were performed for FGFR3-mutated tumors. Treatment effects on cell viability for FGFR3-altered cell lines were evaluated. Synergistic treatment effects were evaluated using the zero interaction potency model.
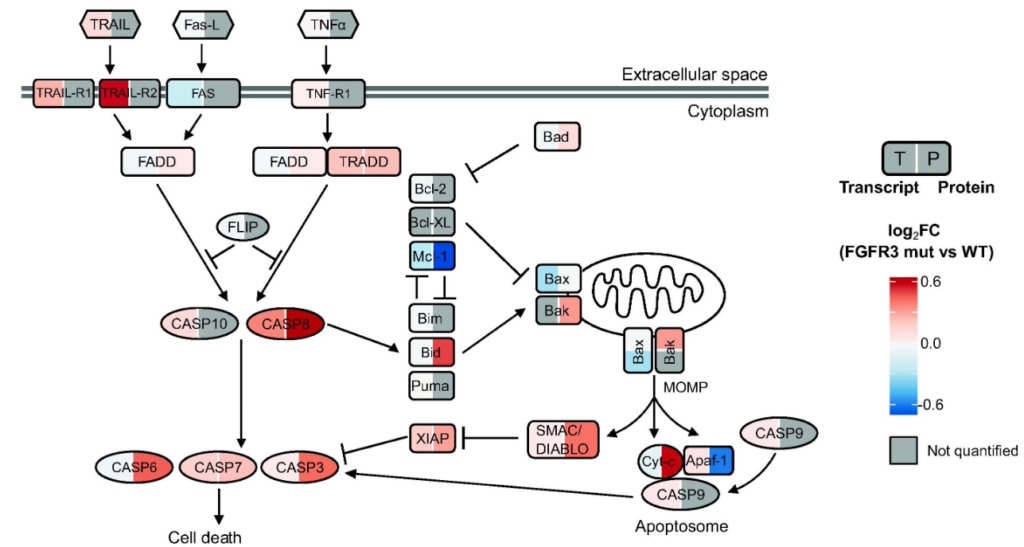
Results and limitations
Five uPGs, covering both NMIBC and MIBC, were identified and bore coarse-grained similarity to transcriptomic subtypes underlying common features of these different entities; uPG-E was associated with the Ta pathway and enriched in FGFR3 mutations. Our analyses also highlighted enrichment of proteins involved in apoptosis in FGFR3-mutated tumors, not captured through transcriptomics. Genetic and pharmacological inhibition demonstrated that FGFR3 activation regulates TRAIL receptor expression and sensitizes cells to TRAIL-mediated apoptosis, further increased by combination with birinapant.
Conclusions
This proteogenomic study provides a comprehensive resource for investigating NMIBC and MIBC heterogeneity and highlights the potential of TRAIL-induced apoptosis as a treatment option for FGFR3-mutated bladder tumors, warranting a clinical investigation.
Patient summary
We integrated proteomics, genomics, and transcriptomics to refine molecular classification of bladder cancer, which, combined with clinical and pathological classification, should lead to more appropriate management of patients. Moreover, we identified new biological processes altered in FGFR3-mutated tumors and showed that inducing apoptosis represents a new potential therapeutic option.
Keywords:
TRAIL; signalling
